You will be told the database and schema names by the DBA, when you are given access.
You will be using your MEB account as authorization to access the data, i.e. you will use the same username and password that you use to log into your computer or VDI account.
If you are running from a Windows machine, the operating system will already have your credentials, and will use those on your behalf.
If you are on a Mac of Linux machine (including matrix and vector), you may have to set up a Kerberos ticket. Do this with kinit, and use the same password you use when logging into the MEB computers (your “Windows password”):
[joelus@matrix ~]$ kinit
Password for joelus@MEB.KI.SE:
[joelus@matrix ~]$If you are using tensor, then this process has been abstracted away into a module:
[joelus@tensor ~]$ ml mebauthNote that when you have been granted a new access, you need to log out and log in again, to use the new access.
Questions? Mail them to dba-support@meb.ki.se. If at all possible, include code and relevant log messages!
SAS
Using CRIME3 as an example dataset, the libname statement looks like this:
libname CRIME3 odbc
noprompt="dsn=kosmos;database=MGRCRIME3"
schema="MGRCRIME3";Note that this example also works unchanged on the compute server (matrix), and with rsubmit.
STATA
Using CRIME3, and the view V_LMED2015 as an example, the load statement looks like this:
odbc load,
table("MGRCRIME3.MGRCRIME3.V_LMED2015")
noquote dsn("kosmos");i.e. you specify the three-level path … The noquote keyword is mandatory.
To access the list of tables and view, use
odbc load,
table("MGRCRIME3.INFORMATION_SCHEMA.TABLES")
noquote dsn("kosmos");Note that these examples works also on the compute server (matrix), unchanged.
R
Using CRIME3 and the V_LMED2015 view as an example, a simple version of R code, using only basic packages, looks like this:
# install the DBI and odbc packages (this is a one-time install)
install.packages(c("DBI", "odbc"))
# load the DBI package (once per session)
library("DBI")
# and possibly also the odbc package ...
# library("odbc")
# connect to the database
dbhandle <- dbConnect(odbc::odbc(), "kosmos")
# make a query
query <- dbSendQuery(dbhandle, 'select ATC, LAKMED from MGRCRIME3.MGRCRIME3.V_LMED2015 where TKOST > 500000')
# get data
result <- dbFetch(query)i.e. you specify the three-level path … but see below for another way of doing this.
To access the list of tables and view, use
# setup as above
column_query <- dbSendQuery(dbhandle,
'select * from MGRCRIME3.INFORMATION_SCHEMA.TABLES')
print(dbFetch(column_query))dplyr also has support for querying the database:
# install DBI, odbc as above
# install dplyr, dbplyr
install.packages(c("dplyr", "dbplyr"))
library(dplyr)
library(dbplyr) ## for in_schema
crime3 <- DBI::dbConnect(odbc::odbc(),
dsn = "kosmos", database = "MGRCRIME3")
lmed15 <- tbl(crime3, in_schema("MGRCRIME3", "V_LMED2015"))
expensive <- lmed15 %>%
filter(tkost > 500000) %>%
select(ATC, LAKMED)Note that these examples works also on the compute server (vector), unchanged.
Python
import pandas as pd
import pyodbc
conn = pyodbc.connect('DSN=kosmos;'
'Database=MGRCRIME3;')
df = pd.read_sql_query('select distinct ATC, LAKMED from MGRCRIME3.V_LMED2015 where TKOST > 500000', conn)
print(df)VS Code
You will need the SQL Server (mssql) extension. You will get a new icon to the left, looking like a server (![]() ). If you click that you will see your connections — with the option to add a connection. Or type F1, then select the MS SQL: Connect command to open the connection workflow.
). If you click that you will see your connections — with the option to add a connection. Or type F1, then select the MS SQL: Connect command to open the connection workflow.
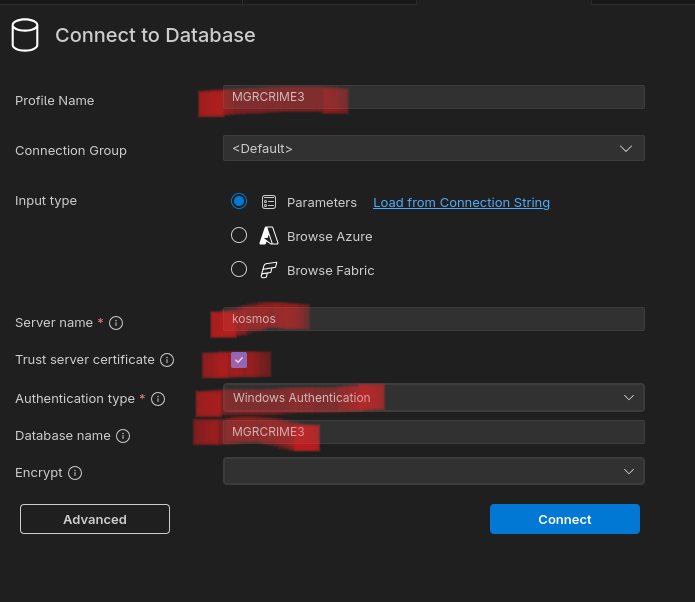
Server name should be kosmos, and you should use Windows Authentication. Check the “Trust server certificate” box. The database name is optional.